Milestone towards a new MIKAIA “AI Authoring 2.0” App
Our new open access journal paper on AI-based Tumor-Stroma Ratio quantification is out!

It is a result of our research towards the AI Authoring 2.0 MIKAIA App, which does pixel-wise segmentation. The current AI Author App does patch-wise classification, which is faster but coarser. In this paper we evaluate
a few-shot based segmentation AI model
(trained to generalize well to other tasks)
and compare it against a “regular” non-few-shot U-Net on the task of tumor-stroma separation.
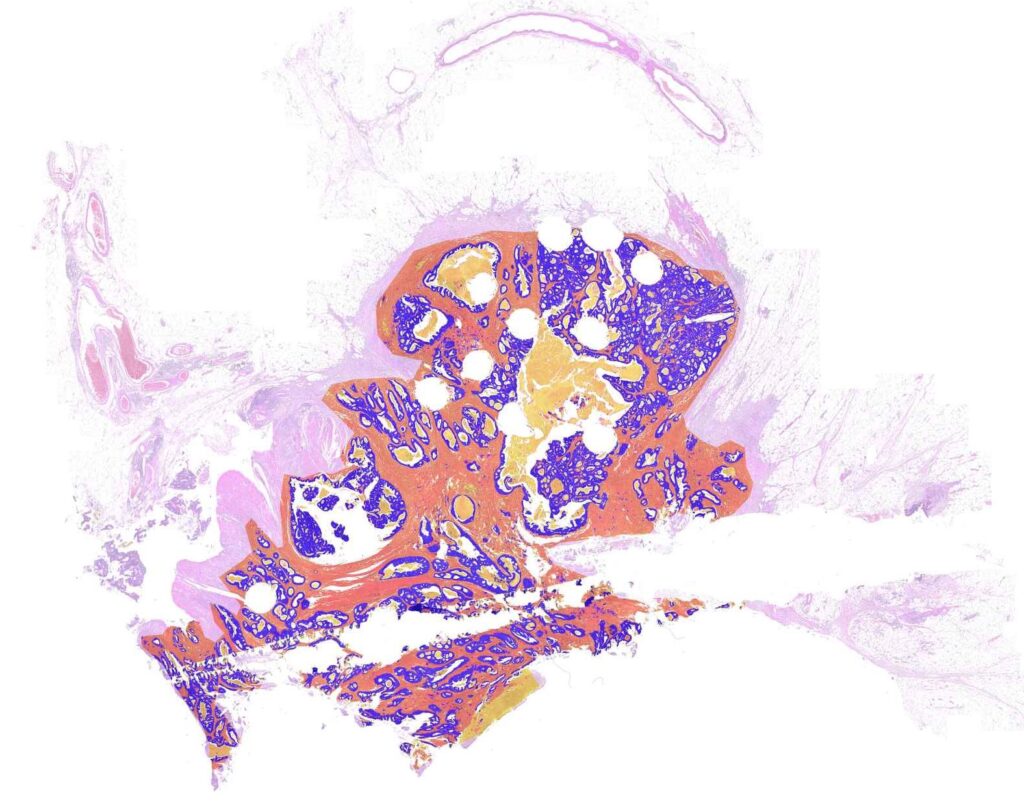
What is the Tumor-Stroma Ratio (TSR) and why is it useful?
A tumor does not only consist of viable tumor cells, but additionally contains stroma, mucus or necrosis. The ratio of the area covered by tumor cells vs. the entire tumor area is denoted the tumor-stroma-ratio.
The tumor-stroma ratio is meaningful in two scenarios:
- Risk stratification: In some cancers, e.g., in colorectal cancer (CRC), the TSR serves as a independent prognostic factor. Counter-intuitively a lower tumor ratio, i.e., a higher stroma ratio, correlates with a poorer prognosis.
“more stroma = higher risk”
Apparently, the tumor’s growth is not hindered by stromal tissue, but the particular tumor seems to be good at simply “consuming” it. - Molecular Pathology: When submitting a sample to Molecular Pathology, e.g. NGS testing, tumor tissue needs to be “scraped” from an area that contains a high ratio of tumor cells, because the scraped cells’ genes or transcripts will be pooled and the NGS result should be dominated by tumor cells and not by other non-tumor cells. Additionally, when interpreting the result, it is helpful to have an accurate estimate of the tumor cells in the tested sample.
A combination of tumor area localization + subsequent TSR quantification would yield an area recommendation system.
An interesting result came out
We asked a panel of pathologists to jude the quality of our AI’s segmentation. An interesting result came out:
The AI segmentation quality looks very good
panel of pathologists
…yet in some cases their own quantitative TSR estimation deviates significantly!?
What is the reason for this seeming contradiction?
The answer is: Tumor-Stroma Ratio quantification actually comprises two tasks:
- Recognizing different tissue types (tumor, necrosis, mucus, …)
- Computing the area ratios
Humans are really good at task 1), but not so good at 2).
Whereas computers (AIs) become increasingly better at 1), and they are perfect at 2).

Read all the details in our open access paper.
The development and experiments were done by our former student research Daniel Firmbach (now at UK Erlangen) under the supervision of our Michaela Benz.
This publication is one of many in the field of Digital Pathology that stems from the collaboration between Fraunhofer IIS and University Hospital Erlangen.




Add comment