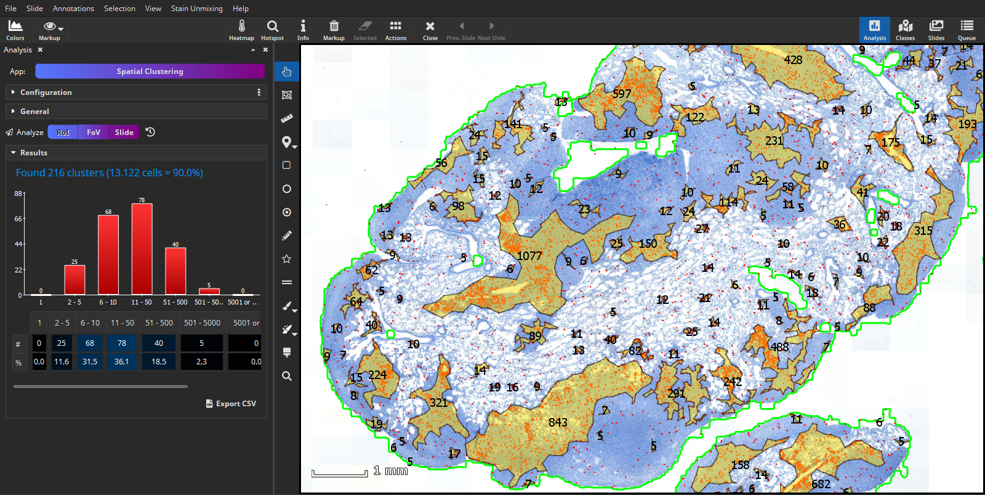
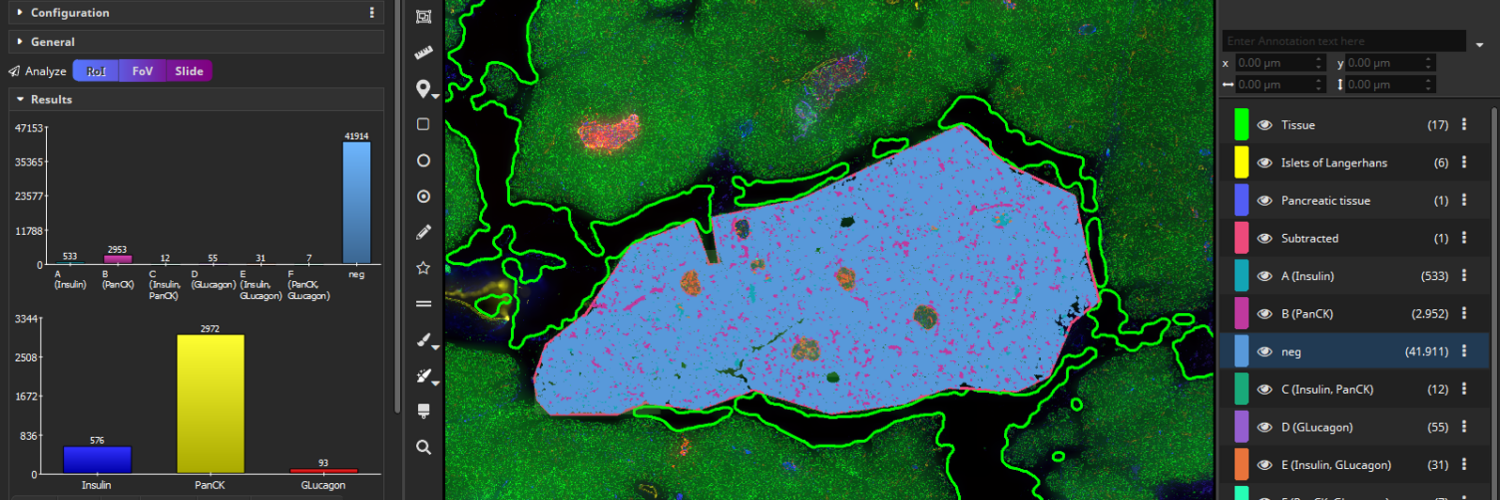
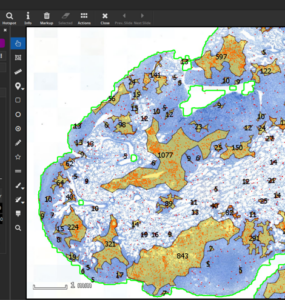
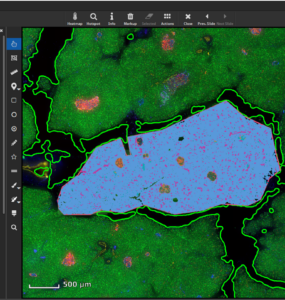
MIKAIA® University is the central MIKAIA® learning portal. Here you can find various App Notes and Video Tutorials that describe typical MICAIA® workflows either from a technical or medical perspective. As we are continuously adding more content...
MIKAIA® University
MIKAIA® University is the central MIKAIA® learning portal. Here you can find various App Notes and Video Tutorials that describe typical MICAIA® workflows either from a technical or medical perspective. As we are...
November 16, 2023