Many recent breakthroughs in cancer therapy fall into the category of personalized medicine. Cancers are characterized more deeply using Molecular Pathology. Another revolutionizing technology that opens up new possibilities to researchers is spatial biology, including its sub-disciplines proteomics and spatial transcriptomics. When reading about these technologies, one is faced with terminology that is far from native to many data scientists who enter this field with a technical background. And that’s where our Molecular Biology 101 for Techies comes in.
To give an example: We are interested in learning how methods such as bulk or single-cell sequencing work. They can sequence the genome, exome, or individual gene transcripts. Along the way, processes such as permeabilization are required and on a microscopic scale, so-called oligos are hybridized to complementary DNA strands and sometimes carry multiple fluorophores that form a unique barcode.
This article will give a brief overview about a range of molecular biology basics. If you are more interested, how these basics are applied in real-world research, read our Spatial Biology 101.
Biological glossary
In the following, we try to collect some of the frequently used vocabulary, and try to organize and explain it without requiring any deep biological prior knowledge.

Let’s begin with a biological glossary of the most relevant basic elements:
| Nucleobases (“bases”) | Nitrogen-containing base DNA strands are made of 4 nucleobases: ACGT (adenine, cytosine, guanine, thymine) RNA strands are made of 4 nucleobases: AGCU (uracil instead of thymine) A DNA molecule is composed of two DNA strands held together by hydrogen bonds between the complementary paired bases. The three-dimensional structure of DNA is a double helix. |
| Nucleotide | Nucleotides are composed of a nucleobase, a five-carbon sugar and one or more phosphate groups. In DNA, the sugar is deoxyribose. In RNA, the sugar is ribose. The abbreviation is “nt”. |
| Oligonucleotide (“oligo”) | Oligonucleotides are sequence of nucleotides, typically created synthetically in a lab. They are used as “probes” for specific DNA or RNA sequences e.g. in DNA microarrays, FISH, Southern blots, PCR. Naming convention: ends with –mer. Examples: 6x nt = Hexamer, 20x nt= 20-mer |
| Gene | Region of DNA that is transcribed as a single protein or a single RNA. Sequence of nucleotides, in the range of 1.000 – 1.000.000 base pairs |
| Genome | Entire set of DNA genes belonging to an organism. Comprises ca 20.000 -25.000 genes |
| Genetics | Study of the genes of an organism on the basis of heredity and variation |
| Epigenetics | Study of phenotypic changes in organisms caused by modification of gene expression |
| Exons | Subset of a DNA sequence that encode RNA. DNA is made up of exons and introns. When it is translated into RNA, only the exons are kept, while introns are discarded. |
| Introns | Subset of a DNA sequence that does not become part of the final RNA product. |
| Exome | Set of all exons, i.e., protein-coding subset of the genome. The whole exome refers to the portion of the human genome that encodes for proteins, known as exons. The exome makes up approximately 1-2 % of the human genome and includes the coding regions of all genes. |
| Transcriptome | All of the RNA molecules that are produced by the cell’s genes. The term “transcriptome” is sometimes used ambiguously: either the entire RNA or only the coding mRNA is meant. · Coding mRNA: 1-4 % of the RNA – codes for proteins · Non-coding: does not give rise to proteins The size of transcriptome varies heavily per cell type |
| Protein | Large molecules comprised of amino acids that perform various functions in the cell. |
| Proteome | All proteins of a cell or a cell compartment that are expressed under precisely defined conditions and at a specific time (~ 1 Mio proteins) |
| Codon | “three-letter word” formed by 3 nucleotides (“bases”) that codes an amino acid, e.g. TTT, ACT, CAG ,… would represent three amino acids. |
| Amino Acids | build blocks for proteins. Proteins are made of only 20 amino acids linked together in chains (“polypeptides”) |
DNA vs. RNA
| DNA | RNA | |
|---|---|---|
| Meaning | “The human code” Gentetic code — the entire genetic information of a organism is stored in the DNA. Nearly all human cells contain the same DNA. So more insightful to look at RNA or proteins. | Responsible for protein synthesis; RNA present in cells varies in quantity and type. |
| Form | Double-helix | Single strand, folded onto itself |
| Stability | Relatively stable | Relatively unstable |
| Nucleobases | 4: A, C, G, T | 4: A, C, G, U |
| Sugar | Deoxyribose | Ribose. Ribose has a hydroxyl group, which makes RNA more chemically labile |
| Subtypes | Protein-coding subset of genome is called exome. | mRNA, tRNA, rRNA, snRNAs, non-coding RNA (“non-coding” means here that this RNA type is not involved in protein synthesis) |
| Processes | Exons are transcribed to mRNA | mRNA delivered to ribosomes (factories in the cytoplasm). Ribosomes read message of mRNA (“recipe”) and assemble amino acids into proteins. |
History excursion: Discovery of the DNA double helix

British physicists and molecular biologists James Watson and Francis Cricks discovered the molecular structure of DNA. For a few years then, the “race for the DNA structure” had been going on, which they won with their publication in Nature on April 25th in 1953. Their paper was only one page long and contained merely 6 references. They combined cues from their own work as well as unpublished literature, notably the famous “photo 51” from their rivaling King’s College London colleague Rosalind Franklin. They had the break-through idea, that the structure must be a double-helix (not a triple-helix as previously falsely postulated by chemist Linus Pauling) where the bases are directed inwards, not outwards. Posed then with the problem that the 4 bases do not have the same sizes and forces and in line with the — at the time well known — observation that two pairs of bases were suspciously always observed at equal amounts, they figured out that not identical but rather complimentary bases interconnected the two leixes (via a hydrogen bond): adenine pairs with thymine, and cytosine pairs with guanine. Watson and Cricks, together with Maurice Wilkins, who pioneered the X-ray diffraction method, later received the Nobel price. Rosalind Franklin, who died only 3 years before from cancer (not unlikely the result of exposion to X-rays in her research), was not honored.
Genotype vs. Phenotype
| Genotype | The genotype refers to the genetic information that an individual inherits from its parents. |
| Phenotype | The phenotype of an organism refers to its observable characteristics or traits, such as physical appearance, behavior, and biochemistry. “phenotype = genotype + environment” |
Omics
By now, a few patterns may have become obvious
- Genomics is the large-scale study of the genome (or part thereof). The genome is the collection of all genes.
- Transcriptomics is the art of measuring the transcriptome (or part thereof), which is the collection of all transcripts that are produced by a cell’s genes.
- Proteomics is the large-scale study of the proteome (or part thereof), which is the collection of all proteins.
| omics | *ome | Unit |
| genomics | genome | gene |
| proteomics | proteome | protein |
| transcriptomics | transcriptome | transcript |
| lipidomics | lipidome | lipids |
| metabolomics | metabolome | metabolites |
Biological processes involved in sequencing
Different technologies can be used for measuring the genome / transcriptome / proteome. These methods involve various biological processes:
| Synthesis | RNA synthesis: catalyzed by an enzyme: RNA polymerase |
| Permeabilization | Making cell membrane permeable so that DNA/RNA can get out |
| Hybridization | Binding of complementary strand to a target strand |
| Replication | DNA is clonally amplified to more DNA, e.g., with PCR Enzyme: DNA polymerase |
| Transcription | DNA is transcribed to mRNA (Process of copying a segment of DNA into RNA) Enzyme: RNA polymerase |
| Translation | RNA is translated to a protein |
PCR
“Thanks” to the Covid pandemic, everyone has heard of PCR. But is a PCR used to measure genes? Not really. It can rather be regarded as a pre-processing step that is required to duplicate the DNA in a small sample to the level that it becomes measurable by sequencing technologies. PCR stands for polymerase chain reaction. The original DNA sample to be copied is called the “template”.
PCR devices perform a sequence of steps in multiple cycles.
- In each cycle, the device first heats up the sample in order to break up the DNA double-helix into two single strands.
- Next, short oligos called “primers” bind to the now single-stranded DNA.
- Then the polymerase enzyme triggers a process called polymerization, in which free-floating T, C, G, or A bases successively bind to one side of the primer. In each turn, only the base that is complementary to the base in the DNA template can attach to the primer and the already bound bases. Base by base, the DNA strands are complemented and new double-helixes are formed.
The steps 1-3 are repeated over and over. In each cycle, the number of DNA strands is approximately doubled.
Sequencing
After the sample has been amplified using PCR, it can be sequenced.
Various types of sequencing exist. The term Next Generation Sequencing (NGS) may be familiar to everyone. But how many generations of sequencing were there before NGS you may ask. Next Gen Sequencing is actually only the 2nd generation, the first one being Sanger Sequencing invented in 1977. It has been used for four decades and was then replaced by NGS, where devices became commercially available in ca. 2005. First generation sequencing suffered from very low throughput. NGS solved this challenge by employing massively parallel sequencing, typically in flow cells. NGS can read hundreds of megabases to gigabases of nucleotide sequence reads in a single instrument run. The market leader for NGS is San Diego based Illumina Inc. Briefly, they use an approach called Sequencing by synthesis, in which a fluorescently labelled base emits light when it is incorporated into a growing DNA. This light is then imaged with a microscope and analyzed.
RNA-Sequencing, abbreviated RNA-seq, refers to the sequencing of RNA instead of DNA. Many devices first reverse-transcribe RNA to “complementary DNA” (cDNA), which is then in turn sequenced with short-read sequencers like Illumina’s HiSeq. A drawback of this approach is that errors are introduced during the reverse transcription. Other devices like NanoString’s nCounter instead measure the RNA directly and do not rely on using cDNA as a proxy for the RNA. So does, real-time RNA sequencing such as the “USB stick” solution by Oxford Nanopore Technologies. In real-time sequencing, bases are reported while the sequencing is still ongoing.
Number of sequenced genes or transcripts
When a technology allows measuring only a limited set of genes, it falls into the category of Targeted Sequencing. Oppositely, when all genes can be distinguished, this is referred to as Whole-Genome-Sequencing.
| Whole-genome sequencing WGS | Method can sequence all genes |
| Whole-exome sequencing WES | Method can sequence all exons (protein coding regiosn of the DNA) |
| Whole-transcriptome sequencing | Method can sequence all transcripts (RNA) |
Sequencing: Bulk → Single Cell → Spatial
Bulk-Sequencing means that the RNA or DNA from many cells is “pooled”. While this sequencing method is simpler, its obvious drawback is that the genes from many cells are measured and so the result may get “polluted” by non-target cells that were merely “bycatch”. For instance in clinical molecular pathology, the intention is usually to sequence only tumor cells in order to identify the tumor subtype and decide which therapy to apply. Cells are “scraped” from the (supposed) tumor area in the tissue and so care has to be taken that this set of cells contains a large portion of tumor cells.
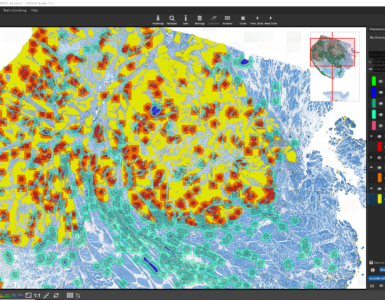
The opposite of bulk sequencing is Single Cell Sequencing, abbreviated “scSeq”. Like in bulk sequencing a pool of cells is first collected, but in a preprocessing step, a device first labels each cell with a unique molecular barcode. The 10x Genomics Chromium Controller, for instance, does this by encapsulating a single cell together with reagents in a droplet and stimulating a micro reaction that leads to the barcoding. Other technologies separate individual cells into micro-wells instead of droplets. After each cell has been barcoded, the regular bulk-sequencing can be carried out. Thanks to the unique barcodes, transcripts that stem from the same cell can be identified and grouped. The next evolution is Spatial Sequencing, where transcripts cannot only be grouped by cell, but where the origin of the cell in the tissue is known. This way, both the morphology and genome can be examined at the same time and the cell’s environment, i.e., its neighbor cells, are also known.
Alternative Sequencing Methods
Fluorescence in situ Hybridization (FISH) is a spatial sequencing technology. Oftentimes, only a very limited (often single-digit) number of genetic events is detected. For instance, FISH HER2/neu assays employ only two probes that visualize HER2 and CEP17 genes, so that their ratio per cell can be calculated in order to determine if a HER2 overexpression exists. If it does, then this is the driver for the tumor. This companion diagnostic test then indicates a targeted therapy with a HER2 inhibitor (“blocker”) such as Trastuzumab.
quantitative PCR (qPCR), sometimes also called real-time PCR, is a variant of PCR that quantitatively reports in real-time the amplification of a DNA molecule. It can be regarded as a targeted sequencing method. The principle of qPCR is that a fluorophore is added to the primer, which is then imaged and measured. The output is the relative gene expression (or mRNA copy number).





Add comment