The TherVacB project, led by Helmholtz Zentrum München, received a five-year funding within the Horizon 2020 program. With TherVacB, a therapeutic vaccine to cure Hepatitis B, the project’s aim is to provide a novel and affordable curative treatment for chronic hepatitis B patients:
The mission of TherVacB is to overcome HBV-specific immune tolerance and to restore antiviral B-cell and T-cell responses by therapeutic vaccination to finally cure HBV.
https://www.thervacb.eu/the-project/
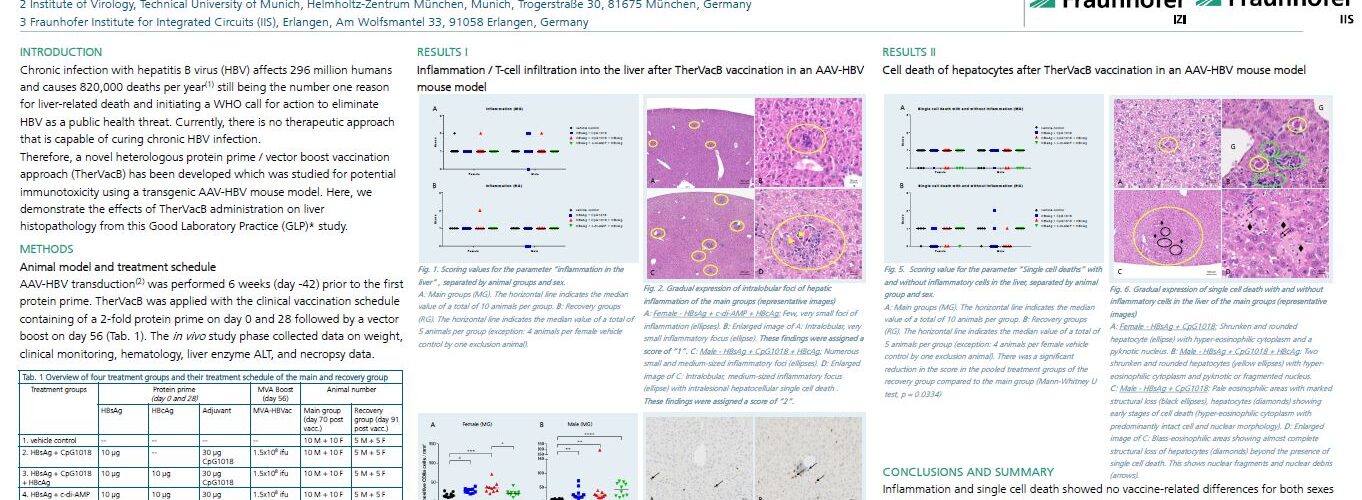
In September 2023, results from a preclinical immunotoxicity study were presented by the project consortium at the 20th European Congress of Toxicologic Pathology (ESTP23) in Basel, Switzerland. Here is the presented poster: Pathohistological evaluation to exclude immunotoxic effects of the TherVacB vaccine in liver tissue – Results from an immunotoxicity GLP study using a transgenic AAV-HBV mouse model.
ESTP23 Poster
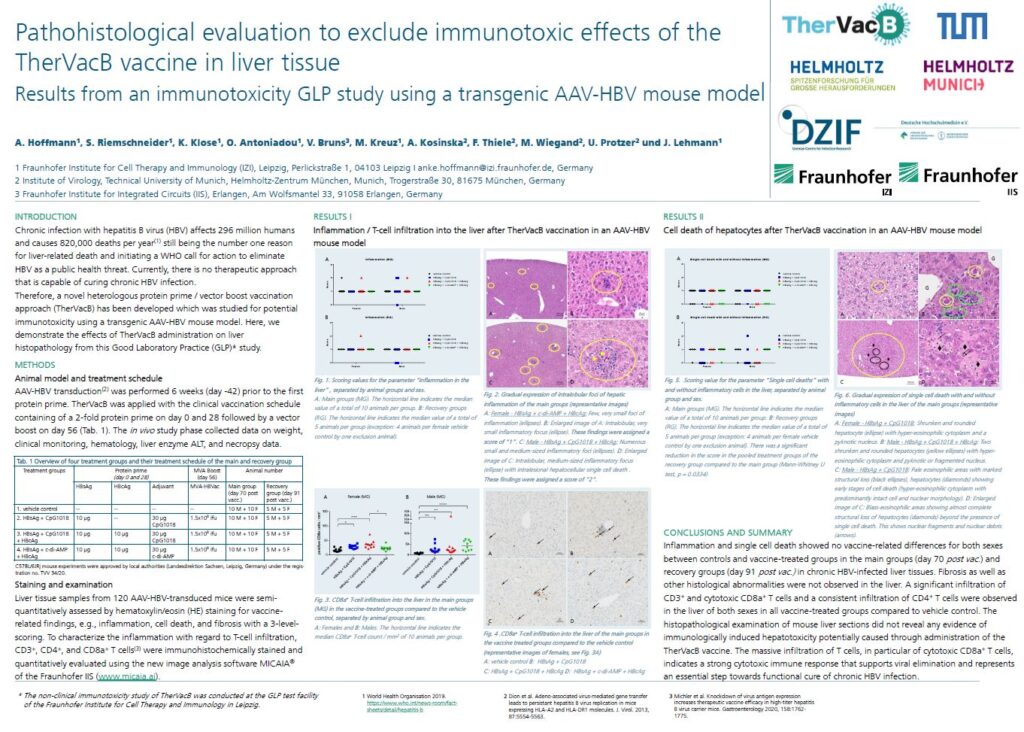
TherVacB vaccine in liver tissue”, © Fraunhofer IZI
Authors
A. Hoffmann1, S. Riemschneider1, K. Klose1, O. Antoniadou1, V. Bruns³, M. Kreuz1, A. Kosinska², F. Thiele², M. Wiegand², U. Protzer², and J. Lehmann1
- 1 Fraunhofer Institute for Cell Therapy and Immunology (IZI)
Leipzig, Perlickstraße 1, 04103 Leipzig - ² Institute of Virology, Technical University of Munich, Helmholtz-Zentrum München
Munich, Trogerstraße 30, 81675 München, Germany - ³ Fraunhofer Institute for Integrated Circuits (IIS)
Erlangen, Am Wolfsmantel 33, 91058 Erlangen, Germany
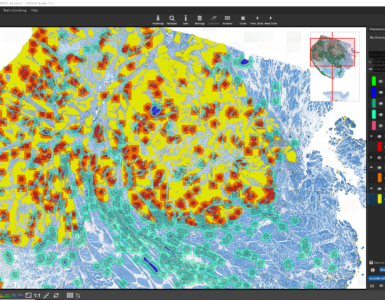
Batch T-cell quantification with MIKAIA
The digital dataset comprised close to 300 whole-slide images (WSI):
- ~100 WSIs each for quantification of CD3+, CD4+ and CD8a+ T cells
- 2 liver sections per scan
- Zeiss CZI format, 40x objective, scanned with Zeiss AxioScan
- 2.5 TB image data in total
The MIKAIA IHC Cell Detection App was used to detect the liver tissue and number of DAB+ cells. The absolute amount and cells/mm² cell density was stated separately per liver section (2 per scan), utilizing the scan region extracted from the Zeiss CZI format. The analysis was performed in three batch jobs – one per marker. Each batch took ~2 hours to compute. The result is a folder each, that contains
- per slide:
- MIKAIA annotation overlay file (*.ano)
- shortcuts to the analyzed files
- low-resolution (~3000x3000px) PNGs with and without burnt-in markup
- CSV with quantitative outputs (area in µm² per tissue, # contained DAB+ cells)
- summary CSV file containing all rows from individual per-slide CSV files
- *.cfg file with used MIKAIA App configuration parameters (loadable into MIKAIA)
- *.micbat MIKAIA batch-analysis file for repeating the batch-analysis on demand




Add comment