Digital pathology and artificial intelligence (AI) go hand in hand to revolutionize medical diagnostics. However, pathology AI models must be able to cope with a very large variety of histological images if they are to be deployed into the field (i.e., “into the wild”). AIs are, therefore, required to be very robust. This robustness in turn is highly dependent on the quality of the underlying datasets that were used to train the AI. Dr. Michaela Benz, chief scientist at Fraunhofer IIS and responsible for the scientific coordination of the development of the image analysis software MIKAIA, and Dr. Petr Kuritcyn, senior scientist and AI expert, explain the importance of robust AI models for digital pathology and the challenges involved.

The use of AI holds great potential for medicine but has not yet become common practice in medical routine. While AI-based image analysis is already standard practice in radiology, microscopy images are still mostly evaluated manually in pathology. What opportunities and possibilities does the use of AI bring to pathology?
Petr Kuritcyn: Currently, digital pathology is still used more often in research than in clinical applications. This will change, however, as pathologists are heavily overworked, and the use of software can help improve the situation. Digital pathologists, for instance, can diagnose while working remotely. Making use of AI based assistance systems also holds great potential when it comes to more trivial tasks such as counting cells. They can very well be taken over by an AI, making daily routines a little bit more efficient. In addition, the results of AI are often very precise and objective, which can further enhance the quality of diagnostics.
Many algorithms and software for this purpose already exist. While the number of AIs that is approved for primary diagnostics for the European market (CE-IVDR) is still small compared to, for instance, radiology, it is growing fast. It’s only a matter of time. However, for AI to be truly implemented in practice, users’ trust in it must be strengthened, and acceptance needs to be fostered. In my view, this is currently happening. On the one hand, because the algorithms are becoming more reliable and achieve very good results, and on the other hand because pathologists are discussing how AI-based methods can be integrated into routine workflows and what benefit these methods can offer.
Can you give us a concrete example for that?
Michaela Benz: A good example from everyday pathology is the characterization of tumors, e.g., Gleason grading for prostate biopsies. This is a very common task that serves as a decision-making basis for therapy selection. Among other things, various so-called biomarkers are used for characterization, which are typically determined by manual evaluation of tissue sections under the microscope by pathologists. Typical tasks in the determination of biomarkers include the counting of certain cell types within a defined area or assessing how strongly a tissue structure has already deviated from its normal healthy layout.
AIs cleared for use in diagnostics today can only do a single and very narrow task. A whole arsenal of AIs is required to assist in the wide variety of tasks pathologists face in their everyday routine.
Dr. Petr Kuritcyn
Another example of a biomarker is the ratio of tumor to stromal area within a tumor. In traditional pathology, the tissue section of a tumor is viewed under a microscope, and pathologists determine the proportion of tumor tissue compared to stroma. This estimation can vary significantly between pathologists and yield subjective results. We have recently shown this in a survey with pathologists from our network. With the help of AI, the assessment of tumor-to-stroma ratio can be automated and performed on a larger scale, allowing for more accurate and efficient analyses.
Another use case where the automatic assessment of the tumor-stroma ratio is helpful is in molecular pathology. Cells are scraped off the tumor tissue for molecular diagnostics. When the DNA or RNA from all scraped cells is pooled, it is important that the sample consists predominantly of tumor cells. Otherwise, the results would be polluted. Here, it is helpful to know the exact ratio.
What considerations should be taken into account while developing an AI-based algorithm for digital pathology?
Petr: Mimicking or even outperforming an experienced pathologists with an AI is not an easy task at all. Therefore, in the first step, we define a clear objective of what exactly the AI should recognize and this determines the AI output. To make it clear: AIs cleared for use in diagnostics today can only do a single and very narrow task, often one type of score for one particular type of staining in one particular organ. A whole arsenal of AIs is required to assist in the wide variety of tasks pathologists face in their everyday routine.
In the second step, we look at the input, i.e., the data that is provided to the algorithm for analysis. In pathology, that’s usually images from biopsies and tissue sections. Based on this, a data processing chain is developed that takes you from input to output. The number of analysis steps and the algorithms required depend on the complexity of the task. The quality of the data we receive from pathologists is crucial for training the AI algorithms. The more accurate these data sets are, the more accurate the results of our algorithms will be.
It’s all about the data, data, data
Petr, you just pointed out that the availability of high-quality datasets is crucial for obtaining robust AI models. What tips and tricks can you share when it comes to creating and annotating data sets?
Petr: Most importantly, data sets need to be representative and cover as closely as possible the heterogenous images the AI will encounter once released into the field. In order to represent this diversity, it is helpful to source samples from multiple centers and digitize the glass slides with different scanners from different vendors. That’s because each lab uses their own staining protocol, and the scanners have different optics, cameras, and also color post processing. In combination these effects lead to surprisingly strong differences in appearance.
Coping with this so called domain shift problem has almost become its own scientific field. Two approaches are either to normalize images at runtime before they are analyzed. Here, color transforming algorithms are available or style-transfer AIs can be used to make the unknown slide look more like the slides from the training set. A drawback is, that this requires additional computational power because one AI is employed to make the “actual” AI work.
Another approach – and we follow this strategy – is to train the AI from the start that it cannot rely on subtle color differences. We achieve this by augmenting our training data set. What we (and many other researchers) do is we duplicate a training image patch and add some random variation to it, e.g. we change the hue or saturation slightly or we unmix the hematoxylin and eosin stains so we can change their intensities separately. You can also simulate camera noise, de-focus, scale or distort, add in JPEG compression artefacts or apply many other augmentation techniques. In the end, the AI has learnt to cope with these variations and will generalize better. To validate this, we use annotated test datasets that really come from different labs and scanners and measure how our AI performs.
It is also critical that the data is annotated as accurately as possible. This means that the structures that the AI is supposed to recognize automatically, must be marked with high precision by experts. Ideally, annotations are created independently by multiple pathologists to avoid individual biases. This enhances the robustness of the algorithm.
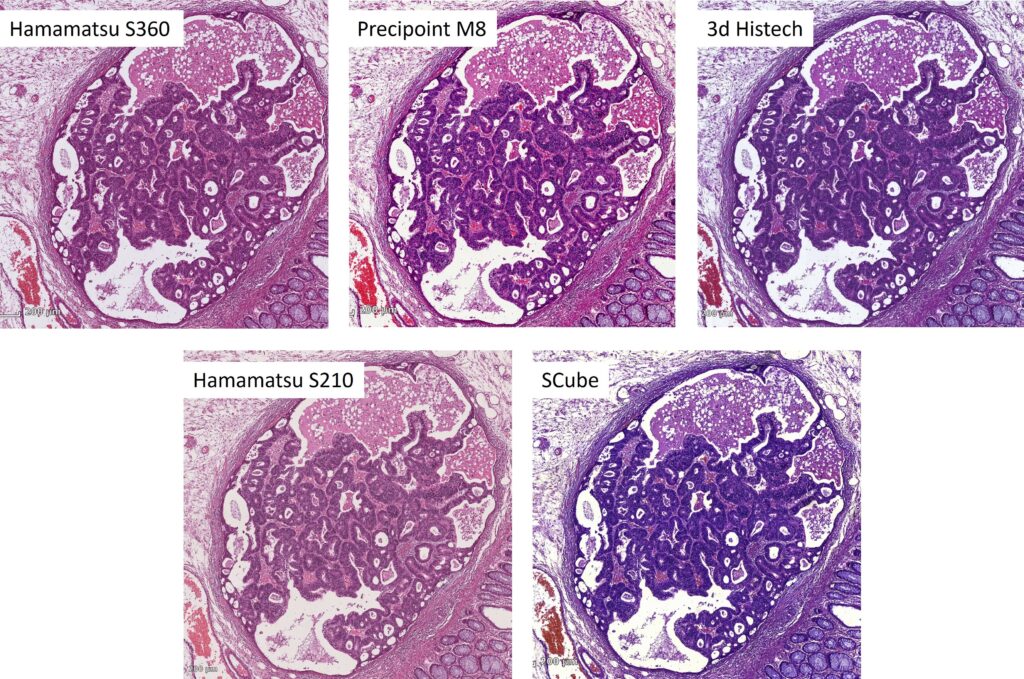
Michaela: It is important that we precisely understand the task and the data. This includes getting an understanding of the variation present in the images. For example, a colon tumor can vary significantly, depending for instance on the subtype or grade, and the AI needs to recognize all these different variations. But large size alone does not guarantee good quality. For example, if a database is built with a million images of the same tumor type, the AI will not work. A good coverage of the entire diversity is more important than the sheer quantity.
How big is big enough?
Can you paint a more detailed picture of just how big these data sets are?
Petr: The size of the database depends very much on the AI approach we use and the complexity of the question at hand. For example, the dataset for an AI that distinguishes multiple tissue types obviously needs to be larger than for a simple decision between A and B. However, there is usually no rule of thumb for the required size, it is based on previous experience. For a complex task, however, we are talking about using approximately one million samples in the dataset. To be clear, by sample we do not mean entire whole-slide-images that are sometimes larger than 100.000 x 100.000 pixels, but we mean individual image patches that we extract from these whole-slide-images.
And how does the training process work exactly?
Michaela: During the development of the software, several data sets – typically a training set, a validation set and a test set – are typically used, each for a different task in the AI development process. These sets contain a large set of examples with their corresponding class labels such as “tumor” or “stroma”. Initially the untrained AI model makes just wild guesses but using the “ground truth” annotations provided by the expert, the model is told whether its guess was right or wrong. Every time it adjusts its internal parameters and after a while it has learnt to identify patterns and relationships.
A large training set alone does not guarantee good quality. If a database is built with a million images of the same tumor type, the AI will still not work well for other types. A good coverage of the entire diversity is more important than the sheer quantity.
Dr. Michaela Benz
For this training procedure the training set is used. The validation set is used to select the best model out of the various models resulting during the training procedure. The test set is initially set aside and not used at all during the training. It is reserved for evaluating on previously unseen data how well (or badly) the fully trained and optimized AI model performs. When a multi-centric data set (meaning data from multiple labs) is available, it is good practice to keep all images from at least one entire lab aside and use them as the test set in order to find out how well the AI generalizes to data from another lab.
What are the particular challenges when developing an AI for pathology? How is this different from other domains such as radiology?
Michaela: There are only few fundamental differences. One difference, however, is the size of images. In pathology, one single digital tissue section can be gigantic. An AI that a dermatologist can use to classify photos of moles operates on images that have a size of 5000 x 5000 pixels at the most, and that’s already quite large. A single whole-slide-image scan, however, can easily be 100 times more or larger. Scans typically have a file size of multiple gigabytes despite employing image compression. The large image size has an impact on the selection of image analysis methods, as standard methods often simply would suffer from a too long computation time.
Pathologists examine a specimen at different magnifications. They require an overview, but for some tasks it is important that they zoom in to the point that individual cells are clearly distinguishable. Some AI approaches mimic this by also analyzing the images at multiple resolutions. Whole-slide-image file formats are built this way, too: They are redundant and not only contain the full resolution images, but also zoomed-down versions, so that a low-resolution overview image is immediately available and does not have to be computed.
Additionally, the diversity of tissue in pathology is particularly challenging. To represent this biological diversity in a meaningful way in a data set is very time-consuming. This diversity is underlined by the fact that pathologists typically select a sub-specialty such as neuropathology, dermatopathology, nephropathology, etc.
In many use cases, it is very challenging to generate objective reference labels for training and also evaluating the AI. For example, objective area estimation is a challenging task for a human. In the above-mentioned study, we were able to demonstrate these varying assessments where both experienced pathologists and our software estimated the area ratio of tumor and stromal tissue. The estimations among the pathologists varied significantly, highlighting the challenge of precisely defining ground truth for data annotation. In another project we considered training an AI to detect if there were inflamed areas that have already progressed into an early stage of a malignant tumor in gastrointestinal biopsies of patient with inflammatory bowel disease. However, we learnt that even the most experienced pathologists frequently disagree in this challenging task, which is why also the medical guidelines require a mandatory second opinion.
Thank you, Michaela and Petr, for sharing all these insights. Let’s talk again some time!
Image copyright (featured image): Fraunhofer IIS / Paul Pulkert




Add comment